Research
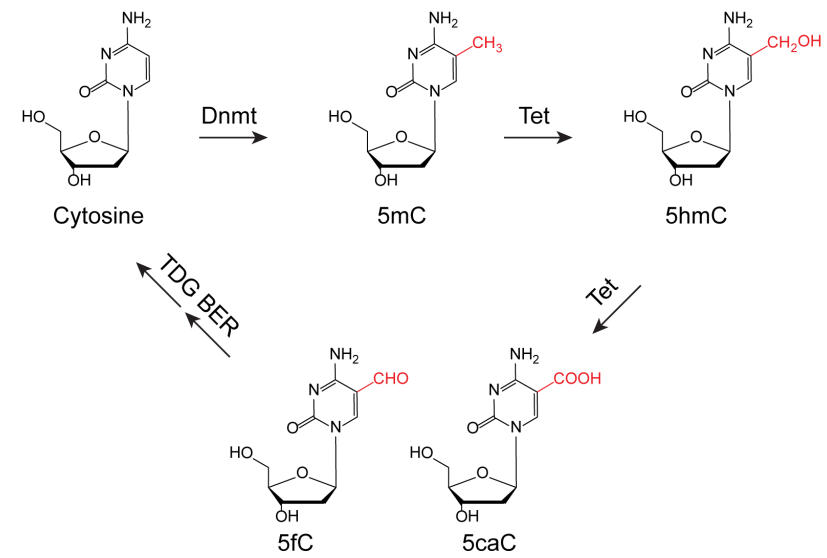
(1) TET and TDG independent mechanisms leading to DNA demethylation
It has been shown that excision of 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by TDG is a downstream event within the active demethylation pathway relying on the TET-catalyzed 5-methylcytosine (5mC) oxidation. Although TDG is required for DNA demethylation in ES cells and for the reprogramming of MEFs into iPSCs, it is dispensable for the demethylation in mouse zygotes (Guo et al., Cell Stem Cell, 2014). Moreover, there is evidence for active DNA demethylation independent of TET3 during early zygotic development (Amouroux et al., Nature Cell Biology, 2016). These observations indicate the presence of other glycosylases or completely unknown mechanisms responsible for DNA demethylation. Identifying enzymes that process oxidized 5mC bases is a priority of our ongoing research.
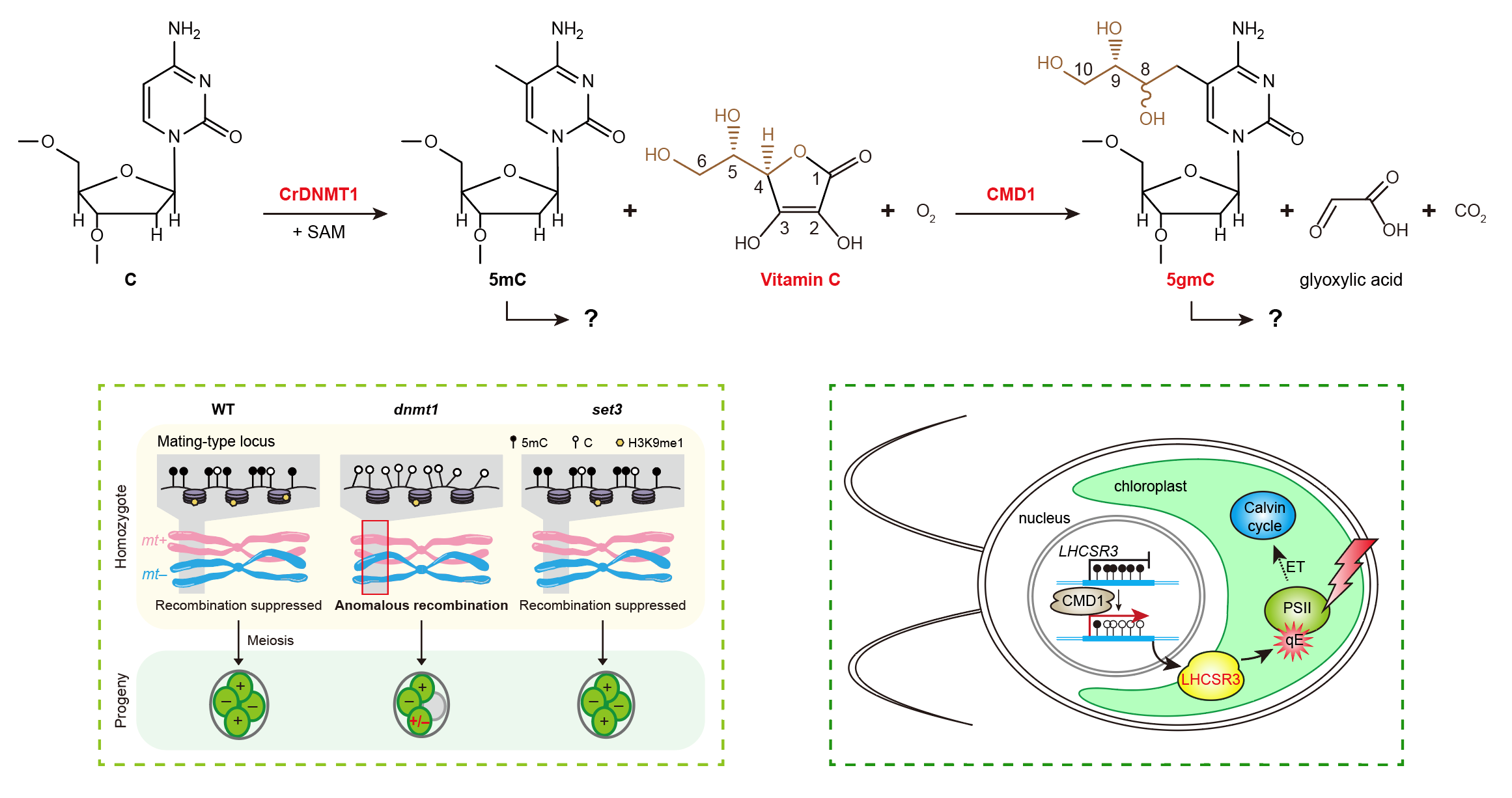
(2) DNA modifications in unicellular eukaryotes
Sequential oxidation of 5mC by TET dioxygenases results in a cascade of additional epigenetic marks and promotes DNA demethylation in mammals (He et al., Science, 2011). However, the enzymatic activity and the function of TET homologs present in diverse eukaryotes remain largely unexplored. Our previous work has showed that CMD1, a TET homolog in the unicellular green alga Chlamydomonas reinhardtii, can catalyze conjugation of a glyceryl moiety onto the methyl group of 5mC and surprisingly utilize L-ascorbic acid (vitamin C, VC) as an essential co-substrate, which functions in the regulation of photosynthesis and epigenetic regulation of environmental adaptation (Xue et al., Nature, 2019). In addition, 5mC plays a vital role in suppressing meiotic recombination at the sex-determining region in Chlamydomonas (Ge et al., submitted). These studies have the potential to reveal new eukaryotic DNA modifications, and their unique biological functions in the epigenetic regulation of environmental adaptation, evolution and so on. The biochemistry and biology of DNA modifications remain rich for exploration.
(3) Epigenetics and gametogenesis
Fertilization provides an opportunity for transmitting not only genetic information but also epigenetic information from parents to early embryos. Oocytes, in particular, exert a profound impact on embryogenesis by transmitting not only a large quantity of RNAs and proteins but also certain epigenetic marks. It has been demonstrated that the maternal epigenome plays vital roles in oogenesis and early embryogenesis. Our recent work has suggested that Tet dioxygenases use an intrinsic auto-regulatory mechanism to regulate their enzymatic activity, thus achieving spatiotemporal specificity of methylome reprogramming, and highlight the importance of methylome integrity for development (Zhang et al., Nature Structural Molecular Biology, 2024). We also focus on two innovative and significant aspects in mammalian male reproductive and developmental biology. 1) We aim to establish immortalized sperm cell line by in vitro culture of round sperm and induction of cell proliferation; 2) Sperm are unique and highly specialized cells with hypermethylated DNA. We are interested in the role of sperm DNA methylation in the development of offspring.
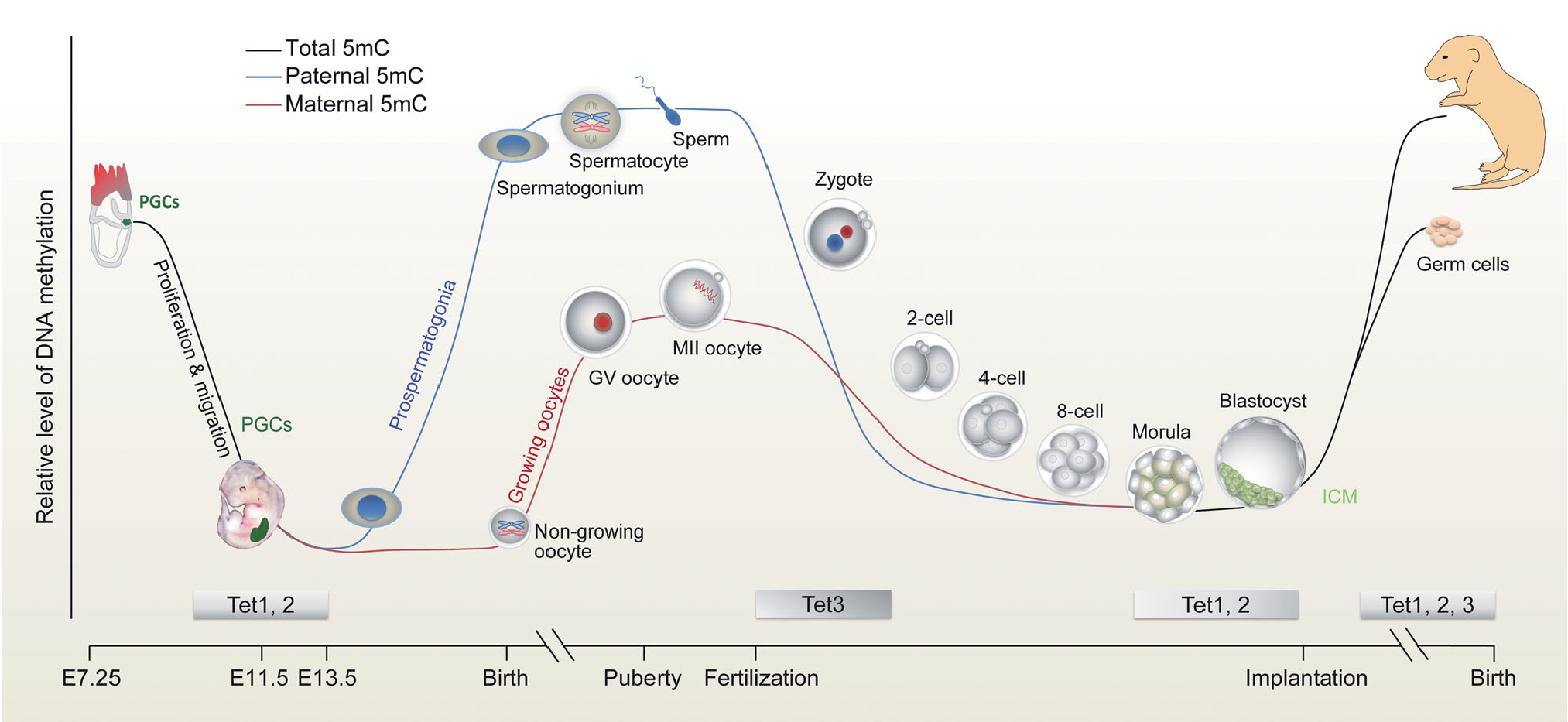
Xu and Wong, National Science Review, 2015
(4) Organ regeneration by blastocyst complementation
Organ shortage is a long-standing worldwide problem. Recently, xenotransplantation of animal organs seems to provide a viable solution to the problem, but immune rejection is an insurmountable chasm. Alternatively, blastocyst complementation, a technique based on injecting wild type pluripotent stem cells into a mutant blastocyst which has developmental defects in a certain organ, has gained popularity in regenerative medicine. Although several studies have successfully demonstrated intra-species and inter-species blastocyst complementation for some organs, including pancreas, thymus, etc., it remains challenging to achieve functional organ complementation between different species owing to developmental incompatibility of tissues from different species. Therefore, we aim to study and overcome these xenogeneic barriers using model systems of rat-mouse blastocyst complementation. These studies will gradually establish the underlying technical framework necessary for the implementation of individualized organ manufacturing.

(5) The mechanism of responding to magnetic fields in eukaryotes
The question of how life responds to magnetic fields is one of the 125 cutting-edge scientific questions published on the 125th anniversary of the founding of Science journal. This topic is considered a frontier field in sensory biology. Despite evidence showing that many organisms utilize magnetic fields for various life activities, key breakthroughs in understanding how they sense these fields are still lacking.
At the macro scale, all species' evolution and life activities occur within the geomagnetic field. While metazoans exhibit diverse responses to environmental stimuli, unicellular protozoa, at the early stages of evolution, may have more direct responses to magnetic fields. Chlamydomonas reinhardtii, a unicellular eukaryote with a billion-year evolutionary history, possesses candidate factors from both animals and plants that may enable it to sense magnetic fields. Therefore, we believe that Chlamydomonas can serve as a valuable model organism for exploring the mechanisms of magnetic induction in eukaryotes. This research may ultimately help us achieve the goal of using artificial magnetic fields to control cellular and even life behavior.
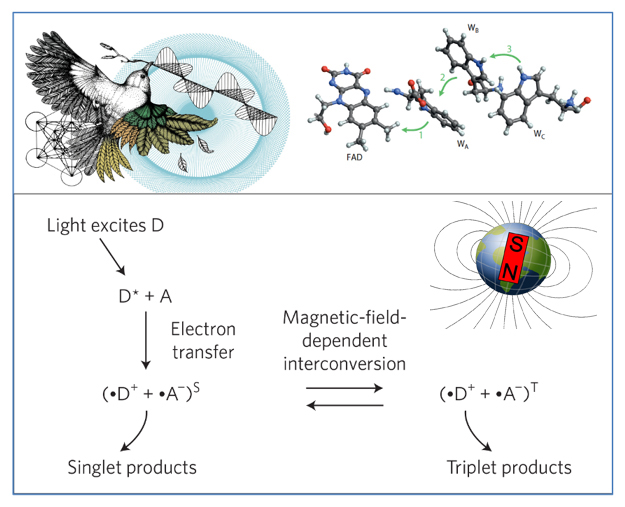
Lohmann et al., Nature Materials, 2016
Recommended articles
- He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303-1307, doi:10.1126/science.1210944 (2011).
- Gu, T. P. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606-610, doi:10.1038/nature10443 (2011).
- Greenberg, M. V. C. & Bourc'his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20, 590-607, doi:10.1038/s41580-019-0159-6 (2019).
- Xu, G. L. & Wong, J. M. Oxidative DNA demethylation mediated by Tet enzymes. National Science Review 2: 318–328 (2015).
- Peters, J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet 15, 517-530, doi:10.1038/nrg3766 (2014).
- Shi, Y., Inoue, H., Wu, J. C. & Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16, 115-130, doi:10.1038/nrd.2016.245 (2017).
- Zhong, C. et al. CRISPR-Cas9-Mediated Genetic Screening in Mice with Haploid Embryonic Stem Cells Carrying a Guide RNA Library. Cell Stem Cell 17, 221-232, doi:10.1016/j.stem.2015.06.005 (2015).
- Hong, T. et al. TET2 modulates spatial relocalization of heterochromatin in aged hematopoietic stem and progenitor cells. Nat Aging 3, 1387-1400, doi:10.1038/s43587-023-00505-y (2023).
- Ko, M. et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 263, 6-21, doi:10.1111/imr.12239 (2015).
- Zheng, C., Ballard, E. B. & Wu, J. The road to generating transplantable organs: from blastocyst complementation to interspecies chimeras. Development 148, doi:10.1242/dev.195792 (2021).
- Bradlaugh, A. A. et al. Essential elements of radical pair magnetosensitivity in Drosophila. Nature 615, 111-116, doi:10.1038/s41586-023-05735-z (2023).
- Komeya, M., Sato, T. & Ogawa, T. In vitro spermatogenesis: A century-long research journey, still half way around. Reprod Med Biol 17, 407-420, doi:10.1002/rmb2.12225 (2018).
- Aranda, S. et al. Thymine DNA glycosylase regulates cell-cycle-driven p53 transcriptional control in pluripotent cells. Mol Cell 83, 2673-2691 e2677, doi:10.1016/j.molcel.2023.07.003 (2023).
- Amouroux, R. et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 18, 225-233, doi:10.1038/ncb3296 (2016).